RNAscope: A Novel in Situ RNA Analysis
In situ analysis of biomarkers is highly desirable in molecular pathology because it allows the examination of biomarker status within the histopathological context of clinical specimens. Immunohistochemistry and DNA in situ hybridization (ISH) are widely used in clinical settings to assess protein and DNA biomarkers, respectively, but clinical use of in situ RNA analysis is rare. This disparity is especially notable when considering the abundance of RNA biomarkers discovered through whole-genome expression profiling. This is largely due to the high degree of technical complexity and insufficient sensitivity and specificity of current RNA ISH techniques.
Here, we describe RNAscope, a novel RNA ISH technology with a unique probe design strategy that allows simultaneous signal amplification and background suppression to achieve single-molecule visualization while preserving tissue morphology. RNAscope is compatible with routine formalin-fixed, paraffin-embedded tissue specimens and can use either conventional chromogenic dyes for bright-field microscopy or fluorescent dyes for multiplex analysis. Unlike grind-and-bind RNA analysis methods such as real-time RT-PCR, RNAscope brings the benefits of in situ analysis to RNA biomarkers and may enable rapid development of RNA ISH-based molecular diagnostic assays.
Biomarkers based on DNA, RNA, and proteins are increasingly used for cancer diagnosis, prognosis, and therapy guidance, heralding the era of personalized medicine.1 RNA biomarkers or gene expression signatures have emerged as a major class of cancer biomarkers, thanks to widespread use of genome-wide gene expression profiling technologies.2, 3 To implement these genomic signatures in clinical diagnostic assays, the current platform of choice is real-time RT-PCR, which is considered the gold standard in gene expression analysis.4 However, this grind-and-bind approach has a serious drawback: the process of RNA extraction destroys the tissue context of gene expression measurements, making it impossible to map the observed signals to individual cells. Furthermore, these assays are prone to interference from unintended cell types (eg, noncancer cells) and from unwanted tissue elements (eg, fibrosis and necrosis). Microdissection techniques can alleviate this problem to some extent,5, 6 but they are too cumbersome and laborious to be useful on a routine basis. By contrast, DNA in situ hybridization (ISH)7 and protein immunohistochemistry (IHC)8 are routinely used in clinical laboratories for DNA and protein biomarker analysis, allowing the integration of molecular information with histopathology for optimal clinical interpretation. To date, use of RNA ISH in clinical diagnostics has been limited to highly expressed genes such as Epstein-Barr virus (EBV)-derived transcripts EBER1/2 in EBV-related diseases.9, 10 Conventional non-radioisotopic RNA in situ hybridization (ISH) techniques lack the sensitivity and specificity required to measure many low-abundance RNA biomarkers reliably.11
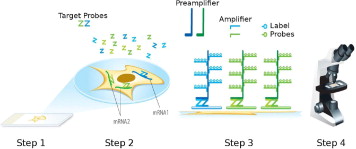
Here, we describe RNAscope, a novel RNA ISH method. Single-molecule visualization in individual cells is achieved through use of a novel probe design strategy and a hybridization-based signal amplification system to simultaneously amplify signals and suppress background. Many of the steps in RNAscope are similar to those in IHC. The RNAscope approach can be used with archival formalin-fixed, paraffin-embedded (FFPE) tissue samples on glass slides, and the stained slides can be visualized under either a standard bright-field microscope (with chromogenic labels) or an epifluorescent microscope (with fluorescent labels). The RNAscope approach allows multiplex detection for up to four target genes (limited by the number of spectrally discernible fluorescent dyes). The ability to analyze gene expression in situ in routine clinical specimen types, as well as high sensitivity and specificity, make RNAscope a promising platform for translating many RNA biomarkers into clinical use.
