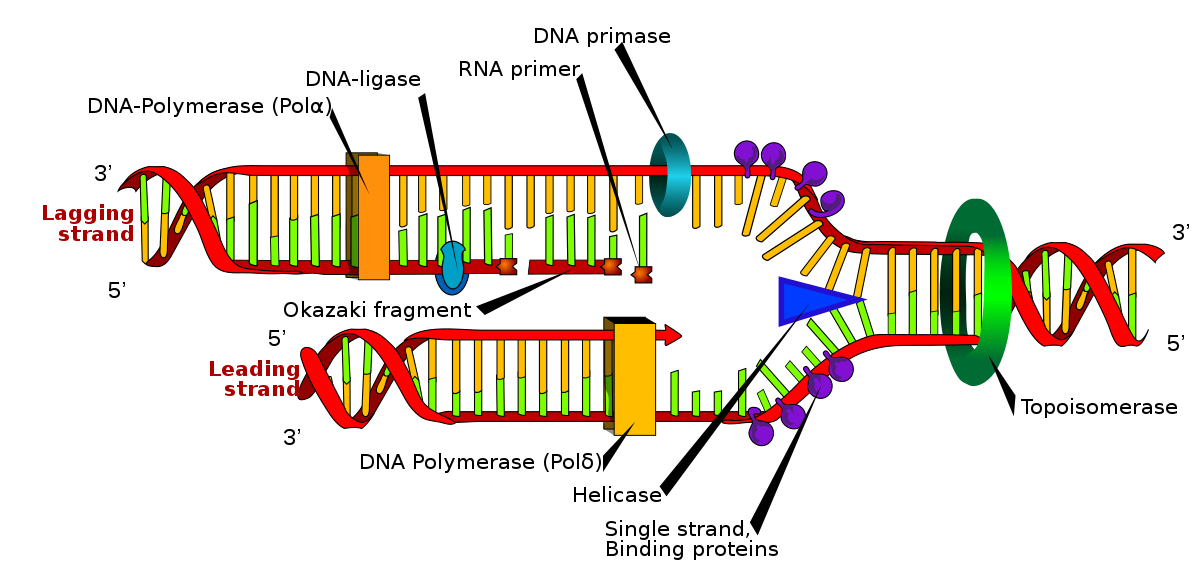
Rna primer
Living organisms use RNA primers to initiate the synthesis of a DNA strand. A class of enzymes called primes add a complementary RNA primer to the de novo reading template on the leading and lagging strands. Starting from the free 3'-OH of the primer, known as the primer terminus, a DNA polymerase can extend a newly synthesized strand. The backbone in DNA replication is synthesized in a continuous piece that moves with the replication fork, requiring only an initial RNA primer to begin synthesis. In the lagging strand, the template DNA runs in the direction 5 ′ → 3 ′. Since DNA polymerase cannot add bases in the 3 ′ → 5 ′ direction complementary to the template strand, DNA is synthesized "backwards" in short fragments that move away from the replication fork, known as Okazaki fragments. Unlike the backbone, this method results in the repeated start and stop of DNA synthesis, requiring multiple RNA primers. Along the DNA template, primase inserts RNA primers that DNA polymerase uses to synthesize DNA in the 5 ′ → 3 ′ direction. [1]
Another example of primers that are used to enable DNA synthesis is reverse transcription. Reverse transcriptase is an enzyme that uses a template strand of RNA to synthesize a complementary strand of DNA. The DNA polymerase component of reverse transcriptase requires an existing 3 'end to begin synthesis. [1]
Primer removal
After insertion of the Okazaki fragments, the RNA primers are removed (the removal mechanism differs between prokaryotes and eukaryotes) and replaced with new deoxyribonucleotides that fill in the gaps where the RNA was present. The DNA ligase then joins the fragmented strands, completing the synthesis of the lagging strand. [1]
In prokaryotes, DNA polymerase I synthesizes the Okazaki fragment until it reaches the above RNA primer. The enzyme then acts simultaneously as a 5 ′ → 3 ′ exonuclease, removing the primer ribonucleotides in front and adding deoxyribonucleotides behind until the region has been replaced by DNA, leaving a small gap in the DNA backbone between the Okazaki fragments. which is sealed with DNA ligase.
In the removal of eukaryotic primers, DNA polymerase δ extends the Okazaki fragment in the direction 5 ′ → 3 ′, and upon finding the RNA primer of the previous Okazaki fragment, it displaces the 5 ′ end of the primer in an RNA flap. single-stranded, which is removed by nuclease cleavage. Cleavage of RNA flaps involves cleavage of flap structure-specific endonuclease 1 (FEN1) from short flaps, or coating of long flaps by replication protein A (RPA) of the binding protein. Single-stranded DNA and sequential cleavage by nuclease Dna2 and FEN1.
